MAPPING QTLíS FOR TRANSFORMATION VARIABLES IN BRASSICA OLERACEA
Penelope A C Sparrow, Philip J Dale, Judith A Irwin.
John Innes Centre, Colney Lane, Norwich, NR4 7UH, UK
ABSTRACT
Previous work over many years has led to the development of protocols to allow selected genotypes to be transformed by Agrobacterium-mediated transformation. These methods may be tailor-made to transform a particular genotype of interest. More often, a genotype amenable to transformation is used and the introduced gene transferred to the line of interest by cross-pollination.† In a winter-sown crop with a vernalisation requirement such as Brassica oleracea, to transfer the desired gene in this way and produce a new variety may take 10-15 years.
Agrobacterium-mediated transformation can be split into three main stages: Agrobacterium infection, selection of transformed cells and shoot regeneration. The aim of this study is to move away from the empirical variation of plant tissue culture methods and to begin to understand the genetic control of three principle factors that affect the transformation process. In this paper, we describe experiments to identify genetic loci associated with shoot and root regeneration.† Work is also ongoing to identify loci associated with background antibiotic resistance (for the selection stage) and tissue sensitivity to Agrobacterium infection.† Preliminary findings will be discussed.
KEYWORDS:† †Regeneration, Tissue Culture, Variation, Genetic Control, Genotype.
INTRODUCTION:
Transformation has been reported for all the main Brassica species, B.rapa (Radke et al. 1992), B. oleracea (De Block et al. 1989, Christey et al. 1997), B. nigra (Gupta et al. 1993), B.juncea (Barfield et al. 1991), B.carinata (Narasimhulu et al. 1992) and B. napus (Moloney et al. 1989).† Protocols vary not only between the Brassica species but also within the species, with some genotypes being particularly recalcitrant to transformation.† Transformation is therefore, highly genotype dependent. Factors that potentially make a genotype amenable to transformation are:
∑ Susceptibility to Agrobacterium: Plant genotypes respond in a diverse way depending on the type and strain of Agrobacterium used (Ramsay and Kumar 1990, Lindsey and Gallois 1990). One reason for this is that some genotypes elicit a hypersensitive response in the presence of Agrobacteria. In extreme cases this can result in necrosis of the tissue at the infection site, hindering transformation events (unpublished data).
∑ Background antibiotic resistance.† The level of antibiotic required to select for transformed cells varies between species.† It is thought that genotypes differ in their natural background resistance to these antibiotics, and hence different levels of selection will be required for different genotypes.† Brassica oleracea is the most sensitive of the diploid Brassicas to amino glycoside antibiotics and variations are also seen within this species (unpublished data).†
∑ Regeneration ability.† Again the ability to regenerate whole plants in vitro is highly genotype dependent, and is therefore likely to be under genetic control.†† Murata and Orton (1987) noted variation in regeneration ability between the Brassica species, with B. oleracea (CC) being the most responsive diploid followed by B. nigra (BB); whilst B. rapa (AA) was fairly recalcitrant to in vitro regeneration.† The interspecific hybrids elicit an intermediate response to that of the parents i.e. AA<AACC<CC.† Variation is also seen within the species Christey et al. (1991) and Irwin et al. (paper 551 of this session, 1999).†††† The genetic control of a number of tissue culture responses has been investigated through quantitative genetic studies, Mendelian genetic analysis and gene mapping (reviewed by Henry et al. 1994).† QTL analysis has progressed further in cereals, such as wheat (Ben Amer et al. 1997) Barley (Mano et al. 1996) and Rice (Taguchi-Shiobara et al. 1997), where a number of QTLís associated to tissue culture response have been identified.† The genetic control of plant regeneration in Brassica is however less clear.† Studies have been carried out into anther culturability (Aslam et al. 1990) where the genetic control was described as Ďextremely complexí.†† Diallel analysis for shoot regeneration in B. napus (Ono et al. 1994) concluded that Ďshoot regeneration ability, was controlled by major gene(s) with an additive and dominant effect, and that these genes could be transferred from high responsive genotypes to low or unresponsive genotypes by sexual crossingí.
It is likely to be a combination of these 3 factors that will determine the transformability of individual genotype.† In this paper we describe a study to investigate the genetic control of shoot and root regeneration in B. oleracea.
MATERIALS AND METHODS:
Plant materials.
A doubled haploid (DH) mapping population derived from a cross between B. oleracea ssp. alboglabra (A12) and B. oleracea ssp. italica †(Green Duke) and a detailed RFLP map associated to this population was supplied by Derek Lydiate for use in this programme. This material was generated by Bohuon (1995) and full map details can be found in Bohuon et al. (1996).
Regeneration procedure.
Preliminary studies were carried out on 30 DH lines from the above-mentioned mapping population.† Mature seeds were surface sterilised and germinated on full strength MS medium supplemented with 3% sucrose and vitamins.†† Cotyledonary petioles and hypocotyl explants were excised from 5-day-old seedlings and cultured on regeneration medium (as above plus 2mg/l BAP).† Regeneration frequencies (number of explants with shoots (or roots) / total number of explants) were scored after 16, 23 and 44 days.† Approximately 60 explants were established per explant type per DH line.
Statistical analysis
Analysis of variance (ANOVAs) for the arcsin ÷ % transformed means was used to detect differences in regeneration frequencies between the doubled haploid lines.†† The MAPMAKERQTL programme (Paterson et al. 1988) was used to detect putative QTLís.
RESULTS AND DISCUSSION:
Variation in both shoot and root regeneration was noted between the two parents and their doubled haploid F1 lines.† The final score data is presented (after 44days).
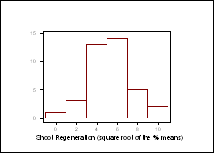
One-way analysis of variance (ANOVA) performed on transformed data (arcsin ÷% means), showed there to be a significant difference in the regeneration responses between the DH lines (p values of less than 0.0001 noted for shoot regeneration from both cotyledonary and hypocotyl explants). The distribution patterns for shoot regeneration, from cotyledonary explants after 44 days in culture, did not approximate to a normal distribution (Figure 1).† A 12x12 diallel is currently being carried out to determine the number of genes associated to this trait.
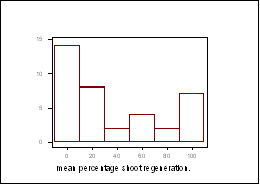
Figure 1: ††††† Frequency distribution for shoot regeneration from cotyledonary explants, † ††††††††††† after 44 days in culture.
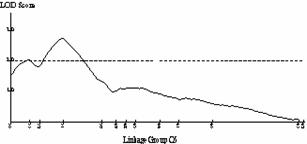
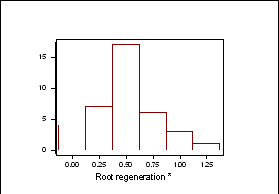
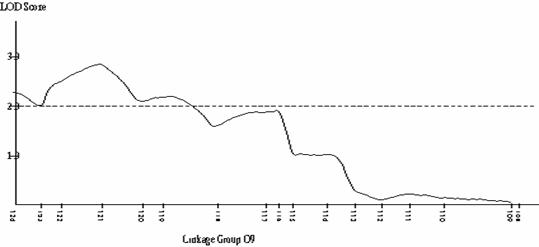
Frequency distributions for shoot regeneration from hypocotyl explants approximated to a normal distribution indicating a polygenic character (Figure 2a).† Shoot regeneration data was entered into MAPMAKERQTL and compared against the B. oleracea linkage map.† Putative QTLís were identified on linkage group O5.† The LOD profile of this QTL can be seen in Figure 2b.
(2a)†††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††††† (2b)