MAPPING RESISTANCE GENES
IN BRASSICA NAPUS
TO LEPTOSPHAERIA MACULANS
S. Roger Rimmer1, M. Hossein Borhan1,2,
Bin Zhu1,2
and Daryl Somers1
1Agriculture and AgriFood Canada, Saskatoon Research Centre, 107 Science Place, Saskatoon, SK
Canada, S7N 0X2
2Department of Plant Science, University of Manitoba, Winnipeg, MB Canada R3T 2N2
ABSTRACT
Black leg,
caused by Leptosphaeria maculans, is one of the more important diseases
of oilseed brassica crops worldwide. Control is generally through the
cultivation of resistant cultivars. Plant breeders have used a number of
sources of resistance genes for this purpose but the relationship of these
genes is not known. To clarify this situation we have done comparative mapping
of resistance to Leptosphaeria maculans (RLM) in populations derived
from crosses of various resistance source parents with Brassica napus cv
Westar as the common susceptible parent. Resistant parents were B. napus
cvs. Cresor, Maluka, and RB87-62. Doubled haploid (DH) mapping populations
were derived from single F1 plants (120-150 DH lines per cross). An
isolate of L. maculans (pl86-12) was used to determine the interaction
phenotype for black leg on both cotyledons and adult plants for all populations
except Cresor H Westar. At least two RLM for PG2 isolates have been previously
mapped on linkage group 6 (LG6) of Brassica napus. RFLP markers, wg8g3,
wg2a11 and tg2b4 from LG6, were linked to a cotyledon resistance gene in Maluka
(cRLMm) when mapped in a population of DH lines from a cross of Maluka H Westar.
Bulked segregant analysis was used to identify AFLP markers linked to cRLMm.
Two AFLP markers were mapped to the interval between the RFLP markers and
RLM. In addition, two RAPD markers BL4 and BL9, linked to a gene conferring
resistance in Cresor at the adult plant stage (aRLMc) were also linked to
cRLMm. Markers linked to cRLMm were also linked to genes associated with
resistance at the cotyledon and adult plant stages in B. napus line
RB87-62, when tested in a DH population of Westar X RB87-62. This information
indicates the importance of LG6 as a source of resistance genes for breeding
programs and genetic studies.
KEYWORD Black leg,
AFLP, RFLP, RAPD, gene cluster
INTRODUCTION
Leptosphaeria
maculans (Desm) Ces. & de Not. is the cause of
black leg of brassica crops. Resistance to the pathogen has been identified
and introduced into breeding lines (e.g. Brassica napus winter rape cv.
Jet Neuf from Europe).
Inheritance of resistance has been reported to be monogenic or polygenic based
on the cultivar and the age of the plant (reviewed by Rimmer and van den Berg,
1992). Genomic location of the some of the resistance genes to L. maculans
has been determined using molecular markers. A locus for resistance to black
leg in B. napus cv. Major was mapped to linkage group six (LG6) using a
population of 105 doubled haploid (DH) lines and L. maculans isolate PHW
1245 (Ferreira et al. 1995). Dion et al. (1995) mapped adult plant resistance
in cv. Cresor. Mayerhofer et al. (1997) mapped a cotyledon resistance gene in
cv. Shiralee, conferring resistance to L. maculans isolates from western
Canada.
Despite genetic
studies on interactions between L. maculans and Brassica spp.,
little is known about the genomic organization of black leg resistance genes.
To determine the possibility of linkage among resistance genes to L.
maculans (RLM) in B. napus cultivars we have mapped RLM genes in
resistant B. napus cvs. Maluka, Cresor and RB 87-62. Here we report the
map location of these genes in DH populations derived from crosses between the
susceptible B. napus cv. Westar with resistant lines of cv. Maluka,
Cresor and RB87-62.
MATERIAL AND METHODS:
Plant Materials:
To produce DH
mapping populations, the susceptible B. napus cv. Westar was crossed
with resistant cvs. Maluka, Cresor and RB87-62. The method of Coventry (1988) was used to produce DH
lines from microspores of F1 plants. Information on the number of
DH lines for each cross and pedigree of the parental lines is presented in
Table 1. DH plants were selfed to produce seeds for pathology tests at the
cotyledon and adult plant stage. Conditions for growing plants were as
described by Keri et al. (1997).
Table 1. Mapping populations developed in crosses
between the B. napus cv Westar (susceptible) and resistant accessions.
Resistant lines Pedigree No
of DH lines
Maluka Haya/Zephyr/Bronowski/3/Chisaya/Zephyr/Bronowski
76
Cresor Canbra X Cresus*3 242
RB87-62 Chikuzen *2/Zephyr X
Bronowski 107
Pathogen inoculation and evaluation of disease
reaction
The interaction
phenotype of the plant response to L. maculans was determined using L.
maculans isolate pl86-12 (pathogenicity group 2 (PG2)). Pycnidiospores
were used as inoculum. Pycnidial inoculum was prepared according to the methods
described in Mengistu et al 1993. Cotyledons of one week old seedling were
inoculated according to the method described by Williams (1985). Interaction phenotype
(IP) of cotyledons were determined after 10 days using a rating scale of 0 to 9
(Williams 1985). Parental lines were used as controls in each test. To
determine the adult plant IP, stem inoculation was carried out at the early
bolting growth stage (GS 3 to 3.2; Harper and Berkenkamp, 1975). A 10 ml droplet of
pycnidiospore inoculum was injected into the stem between the second and the
third node using a hypodermic needle. Adult plant IP was based on the
percentage of discoloration on a section of the stem 5 mm above the inoculation
point and on a scale of 0 to 5 (0 resistant and 5 fully susceptible). The
methodology for evaluation of the adult plant interaction phenotype for 242
Cresor ´ Westar derived DH lines was described by Dion et al. (1995).
DNA extraction and Southern hybridization
Leaves from 6-8
weeks old plants were harvested and freeze dried. Freeze dried tissue (0.5 g )
was used for DNA extraction according to the method described by Sharp et al.
(1995). DNA was blotted onto N+ nylon membranes according to the instructions
of the manufacturer (Amersham). Hybridization was carried out as described by
Sharp et al. (1995).
RFLP, AFLP and RAPD
Restriction
fragment length polymorphism (RFLP) markers were provided by TC Osborn, University of Wisconsin, Madison, USA. Polymorphism was detected by
digesting DNA with EcoRI or HindIII. DNA inserts of RFLP probes were amplified
by polymerase chain reaction (PCR) as described by Ferreira et al. (1994).
Amplified DNA was radio-labeled with [32P] by random priming (Feinberg
and Vogelstein 1983). Amplified fragment length polymorphism (AFLP) was
carried out on EcoRI and MseI digested DNA essentially as described by Vos et
al. (1995). Linked AFLP markers were first identified by bulked segregant
analysis (BSA) (Michelmore et al. 1991) then mapped to a population of
resistance and susceptible DH lines. Randomly amplified DNA polymorphism
(RAPD) markers were generated with primers obtained from the University of British
Columbia, Canada. The PCR reaction contained 10 ng DNA, 0.5 U Taq DNA polymerase
(BRL, Mississauga, Canada), 50 mM KCl, 2.5 mM MgCl2
, 200 μM of each dNTP and 0.2 μM primer. Amplification of DNA was
carried out on a PCT-200 DNA Engine thermocycler (MJ Research, INC. U.S.A).
The cycle parameters were 95 1C -1:30 min (1
cycle); 95 1C- 20s, 36 1C -1 min, 72 1C -1 min (35 cycles); 72 1C B 7 min (1
cycle). PCR products were separated on a 2 % (w/v) agarose gel in 1 X TAE by
electrophoresis at 100 V for 3 h. Gels were stained in ethidium bromide and
photographed on a digital gel-documentation system.
Linkage analysis
Linkage analysis
and mapping of molecular markers were carried out using MAPMAKER/EXP 3.0
(Lander et al. 1987) with the minimum LOD score of 3.0 and a maximum
recombination fraction of 0.3. Interaction phenotype was mapped as a Mendelian
trait in each DH population.
RESULTS
Mapping of RLM
loci was initiated by identifying molecular markers linked to the cRLM in
Maluka (cRLMm). BSA was used with 30 DH lines to obtain AFLP markers linked to
cRLMm. Of 84 primer combinations tested, 11 primer combinations gave markers
linked to the cRLMm. These were mapped on the 30 individual DH lines and 5
markers were linked to the resistance locus. Two AFLP markers, which
co-segregated with cRLMm, were mapped using 48 additional DH lines and
recombinant families were identified. In order to determine the genomic
location of cLMRm, RFLP markers were used. RFLP markers on LG6 of B. napus
map (Ferreira et al 1994) were used since LG6 contained L. maculans
resistance loci (Ferreira et al.1995; Mayerhofer et al. 1997). Of all the RFLP
markers from LG6 mapped on 76 DH lines, three (wg8g3, tg2b4 and wg2a11) were
linked to cLMRm (Figure 1).
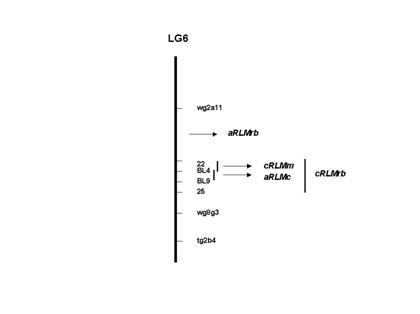
Figure 1. Position of resistance genes to Leptosphaeria
maculans in cultivars Maluka, RB87-62 and Cresor in linkage group 6 (LG6)
of Brassica napus map. wg8g3, tg2b4 and wg2a11 are RFLP markers
developed by Ferreira et al. (1994). BL4 and BL9 are RAPD markers. 22 and 25
are AFLP markers. Loci named as cRLMm and cRLMrb confer resistance to L.
maculans at the cotyledon stage in cvs. Maluka and RB87-62 respectively.
Loci for adult plant resistance in cultivar Cresor and RB87-62 are indicated as
aRLMc and aRLMrb respectively. The figure is not to scale.
The adult plant
resistance gene in Cresor (aRLMc) was tagged using RAPD markers. A population
of 242 DH lines from a cross of Westar ´ Cresor was used for
mapping. Two RAPD markers (BL4, BL9) flanked the aRLMc locus. BL9 was closer
to aRLMc with 3 recombinant families and 6 recombinant families for BL4.
These RAPD markers when applied to DH lines of Westar ´ Maluka population provided
evidence of linkage between cRMLm and aRLMc. BL4 and BL9 co-segregated with
resistance and showed 1 recombinant DH line out of 76 DH lines tested. The
markers linked to aRLMc and cRLMm were then used to map cotyledon and adult
plant resistance genes in RB87-62 (cRLMrb, aRLMrb). RFLP and AFLP markers
linked to cRLMm locus were also linked to cRLMrb when mapped on 30 DH lines of
Westar ´ RB87-62 cross. The RAPD markers BL4 and BL9 were mapped on 107 DH
lines of Westar ´ RB87-62 cross. BL4 and BL9 co-segregated but two recombinant
families were detected between these markers and cRLMrb. The IP of RB87-62 for
adult plant resistance to L. maculans were tested in the green house.
Comparison of these data for 107 DH lines with cotyledon resistance of the same
DH lines showed that aRLMrb is linked to cRLMrb and only 7 DH lines showed
recombination between these two loci. The position of resistance genes in
cultivars Maluka, Cresor and RB87-62 are shown in Figure 1.
DISCUSSION
In cvs. Cresor
and Maluka, resistance is controlled by a single locus as indicated by
segregation ratio of 1 resistance : 1 susceptible in DH lines (for aRLMc: X2:
5.19, P:0.01-0.025; for cRLMm X2: 1.37, P:
0.1-0.25). For RB87-62 segregation for resistance in F2 individuals
indicated that resistance, both at the cotyledon stage and for adult plants, is
monogenic. However segregation distortion occurred in the DH lines of this
cross as more than 80 % of the lines were susceptible when tested at the cotyledon
or adult plant stage. Segregation distortion has been reported for Brassica
populations developed by microspore culture (Ferreira et al. 1994; Mayerhofer
et al. 1997).
The resistance
genes in the three accessions of B. napus that we studied are all
located in LG6 of the B. napus map developed by Ferreira et al. (1994).
At least two other B. napus resistance genes to L. maculans have
been located to LG6. Ferreira et al (1995) identified a major locus (LEM1)
controlling cotyledon resistance to isolate PHW 1245 of L. maculans in B.
napus cv. Major. This gene was located between RFLP marker wg2a3b and
tg5d9b on LG6 south of the cRLMm loci mapped in the present study. A cotyledon
resistance gene in B. napus cv. Shiralee (LmR1) to a PG2 isolate of L.
maculans also mapped on LG6. Since the markers used by Mayerhofer et al.
(1997) for mapping LmR1 are different from ours, it is not possible to
determine the position of resistance loci identified in our study with respect
to the LmR1 locus. However, considering the pedigrees of Maluka (Haya //
Zephyr/Bronowski/3/Chisaya // Zephyr/Bronowski) and Shiralee (Haya //
Zephyr/Bronowski/3/RC33 // BJ168/Cresus-o-Precose) it is likely that LmR1 and
the cotyledon resistance gene in Maluka (cRLMm) are allelic as proposed by
Mayerhofer et al. (1997).
It is also
probable that cRLMm and cRLMrb loci are allelic as Maluka and RB87-62 share
common parents in their breeding pedigrees (Table 1). Molecular markers used
for mapping cRLMm and cRLMrb showed the same polymorphic patterns in both
populations indicating a high level of similarity for this part of the genome
between Maluka and RB87-62. However, we consider the two cotyledon resistance
alleles in Maluka and RB87-62 to be linked but not allelic (Figure 1). This is
due to the position of AFLP markers which flank cRLMm but are located on one
side of the cRLMrb locus. To resolve the map positions of cRLMm and cRLMrb we
are continuing mapping of markers linked to these loci with a larger
population. The adult plant resistance gene in cultivar RB87-62 (aRLMrb) is
linked to the cotyledon resistance locus in this cultivar (cRLMrb) but not
allelic as 7 recombinant DH families were detected between these two loci.
The adult plant
resistance gene in cultivar Cresor (aRLMc) is linked to cRLMm and cRLMrb but
not allelic to these loci. This is evident from the position of two RAPD
markers BL4 and BL9, which flank aRLMc but map to one side of cRLMm and cRLMrb
(Fig 1). Except for RFLP marker wg2a11, most of the markers linked to RLM
genes in LG6 are located south of RLM genes (Figure 1). RFLP probe wg2a11 is
distantly (~ 24 cM) linked to the RLM genes. Difficulty with identifying
markers from this interval of LG6 has also been encountered by Mayerhofer et
al. (1997) for mapping LmR1. They proposed that this might be because LmR1 is
located toward the end of LG6.
ACKNOWLEDGEMENTS
This work was
supported in part by grants to SRR from the Western Grains Research Foundation,
Agriculture and Agri-Food Canada and the Natural Sciences and
Engineering Research Council of Canada. The technical support of Ms P. Parks
is gratefully acknowledged.
REFERENCES
Coventry, J., Kott, L.,
Beversdorf, W. D. (1988). Manual for microspore culture technique for Brassica
napus. Ontario Agriculture publication No. 0489. The University of Guleph, Guelph, Ontario, Canada.
Dion, Y., Gugel, R. K., Rakow, G. F. W.,
Séguin-Swartz, G., Landry, B. S. (1995). RFLP mapping of resistance to the
blackleg disease [causal agent, Leptosphaeria maculans (Desm.) Ces. et
de Not.] in canola (Brassica napus L.). Theor. App. Gen. 91: 1190-1194.
Feinberg, A. P., Vogelstein, B. (1983). A technique
for radiolabelling DNA restriction fragments to a high specific activity. Ann
Biochem. 132: 6-13.
Ferreira, M. E., Rimmer S. R., Williams, P. H.,
Osborn, T. C. (1995). Mapping loci controlling Brassica napus to Leptosphaeria
maculans under different screening conditions. Phytopathology, 85: 213-217.
Ferreira, M. E., Williams, P. H., Osborn, T. C.
(1994). RFLP mapping of Brassica napus using doubled haploid lines.
Theoretical and Applied Genetics, 89: 615-621.
Harper, F. R. and Berkenkamp, B. (1975). Revised
growth-stage key for Brassica campestris and B. napus. Can. J. of Plant Sci. 55: 657-658.
Keri, M., van den Berg, C. G. J., McVetty, P. B. E.,
Rimmer, S. R. (1997). Inheritance of resistance to Leptosphaeria maculans
in Brassica juncea. Phytopathology, 87: 594-598.
Lander, E. S., Green, P., Abrahamson, J., Barlow, A.,
Lincoln, S. E., Newburg, L. (1987). MAPMAKER: An interactive package for
constructing primary genetic linkage maps of experimental and natural
populations. Genomics, 1: 174-181.
Mayerhofer, R., Bansal, V. K., Thiagarajah, M. R.,
Stringam, G. R., Good, A. G. (1997). Molecular mapping of resistance to Leptosphaeria
maculans in Australian cultivars of Brassica napus. Genome, 40:
294-301.
Michelmore, R. W., Paran, I., Kesseli, R. V. (1991). Identification of markers linked to disease
resistance genes by bulked segregant analysis: A rapid method to detect markers
in specific genomic regions by using segregating populations. Proc. Nat. Acad.
Sci. 88: 9828-9832.
Rimmer, S. R., and van den Berg, C. G. J. (1992).
Resistance to oilseed Brassica spp. to blackleg caused by Leptosphaeria
maculans. Can. J. Plant
Pathol. 14: 56-66.
Roy, N.N. 1984. Interspecific
transfer of Brassica juncea-type high blackleg resistance to Brassica
napus. Euphytica 33:295-303
Sharp, A. G., Parkin, I. A. P., Keith, D. J., Lydiate, D. J. (1995). Frequent nonreciprocal
translocations in the amphidiploid genome of oilseed rape (Brassica napus).
Genome, 38: 1112-1121.
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de
Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., Zabeau,
M. (1995). AFLP: a new technique for DNA fingerprinting. Nuc. Acid Res., 23:
4407-4414.
Williams, P. H. (1985). Crucifer Genetics Cooperative
(GrGC) Resources Book, Department of Plant Pathology, University of Wisconsin, Madison, WI.
