GENETIC MANIPULATION FOR ALTERED OIL QUALITY IN BRASSICAS
Stoutjesdijk P.A, Hurlstone C, Singh S.P, Green A.G.
CSIRO Division of Plant Industry, Canberra, ACT, Australia
ABSTRACT
Genetic engineering methods are being used to modify the fatty acid profile of elite Australian germplasm of Brassica napus and B. juncea. Cosuppression vectors carrying oleate desaturase genes from each species have been constructed and transferred into Australian elite breeding lines of B. napus and B. juncea using Agrobacterium tumifaciens plant transformation techniques. Modifications to existing Brassica transformation protocols and the use of a intron-interrupted hygromycin-resistance gene as the selectable marker have resulted in improved transformation efficiencies. Silencing of the endogenous oleate desaturase genes have resulted in substantial increases in oleic acid levels, up to 89% in B. napus and 73% in B. juncea.
KEYWORDS: Canola, Indian mustard, oleic acid, desaturation, cosuppression
INTRODUCTION
The Australian canola industry has grown rapidly in recent years and has considerable scope for further expansion to meet a growing domestic utilisation of canola oil as well as a large world trade in canola seed. Two factors will be important driving forces of expansion.
Firstly, there are increased domestic and export market opportunities for canola oil that can be realised through the development of high-oleic canola to replace saturated palm oil in food service applications. High-oleic oils have been shown to have equivalent heat stability to saturated fats and are therefore suitable replacements for them in commercial food-service applications that require long-life stability. Additionally, high-oleic oils are more nutritionally beneficial because oleic acid has cholesterol-lowering properties, whereas saturated fatty acids tend to raise blood cholesterol levels. The high-oleic type is expected to become a significant proportion of the international canola market in the near future as a result of development of this quality type in other canola producing and exporting countries. To capitalise fully on this opportunity requires the development of Australian B. napus canola varieties in which oleic acid content is raised from its current level of around 60-65% to as close to 90% as possible.
Secondly, there are agricultural opportunities to increase canola production by expanding into the drier regions of the Australian cereal belt. This is being addressed through both the development of earlier-maturing varieties of B. napus canola and the introduction of B. juncea as an alternative source of canola quality oil. Current double-low germplasm of B. juncea has around 45% oleic acid, significantly below the level present in B. napus and B. rapa, and insufficient for the oil to meet market specifications for canola oil. For B. juncea to become a viable alternative source of canola oil, oleic acid must be raised to at least 60%. The opportunity also exists to develop very high-oleic (90%) B. juncea.
Increases in oleic acid content can potentially be achieved by reducing the activity of oleate desaturase (oleoyl-PC D12-desaturase) the enzyme which converts oleate into linoleate in the developing seed. EMS mutants have been successfully obtained which raise oleic acid up to 80% (Auld et al., 1992), however mutants having further increases above this level have been associated with undesirable agronomic effects (Kinney, 1994). It is considered likely that there are several genes coding for the D12-desaturase enzyme in B. napus seed, and that some of these genes also encode production of D12-desaturase in vegetative tissue. Mutations in these latter constitutively-expressed genes can have detrimental effects on vegetative tissue where the correct fatty acid composition is required for normal membrane structure and function. Molecular mechanisms of inactivating genes, such as antisense and cosuppression, can be implemented in a tissue-specific manner enabling the inactivation of all copies of D12-desaturase in the developing seed without affecting gene expression in other tissues. Antisense and cosuppression methods have already been successfully employed to raise oleic acid up to 85% in both B. napus (Kinney, 1994) and soybean (Kinney, 1997).
In order to produce high-oleic quality in Australian Brassica germplasm we have implemented a D12-desaturase gene cosuppression strategy. This paper describes the progress made in achieving transformation in elite Australian breeding lines of B. napus and double-low B. juncea and in raising oleic acid content in both species.
MATERIAL AND METHODS
Plant material
The advanced breeding lines of Brassica napus RI25 and BLN1239 were provided by Agriculture Victoria and NSW Agriculture respectively. A zero-erucic acid, low-glucosinolate line of Brassica juncea (815-1-6-2) was provided by CSIRO Plant Industry.
Binary vectors
The oleate desaturase gene from B. juncea had previously been cloned (Singh et al., 1995) and the corresponding gene from B. napus was cloned by RT-PCR. Cosuppression constructs were made by positioning either the B. napus or B. juncea D12-desaturase coding region in sense orientation downstream from a truncated form of the napin seed storage promoter (FP1). These constructs were cloned blunt-ended into the Not1 site of the p35SH-iC binary vector previously described by Wang et al. (1997) as shown in Figure 1. An intron-interrupted hygromycin resistance gene (Hph) driven by the 35s promoter was used as the selectable marker (Wang et al., 1997). Binary vectors containing the appropriate cosuppression constructs were transferred to Agrobacterium tumifaciens (AGL1) and used in transformation experiments with B. napus and B. juncea according to the transformation protocols described below.
Transformation methods
The transformation protocol follows that of Bade and Damm (1995) with minor modifications. Etiolated hypocotyls were cultured in liquid media for one day to induce callusing prior to being inoculated with Agrobacterium. Following a one day co-cultivation period the explants were plated onto shoot induction medium (SIM, MS based) containing hygromycin and a two-step selection was employed. The first two weeks of selection for transformants was performed on SIM media containing 5 mg/l and 10 mg/l hygromycin for B. napus lines and B. juncea respectively. Further selection was performed on 20 mg/l hygromycin for B. napus and 30 mg/l hygromycin for B. juncea. Green plantlets were transferred to root induction media (RIM) containing lower levels of hygromycin, 10 mg/l and 15 mg/l for B. napus and B. juncea respectively. A selection of putative transformed B. napus plants carrying the D12-desaturase cosuppression constructs were analysed by standard Southern analysis techniques. Twenty out of 21 plants analysed were positive for the presence of the hygromycin gene.
A.
 |
B.
 |
Figure 1. A. Diagram of cosuppression constructs used to silence endogenous D12 desaturase activity in B. napus and B. juncea. B. Diagram of p35SH-iC used to transform B. napus and B. juncea using the hypocotyl transformation protocol of Bade and Damm (1995). Cosuppression constructs were cloned blunt ended into the blunt-ended NotI site shown above.
RESULTS
Preliminary experiments using binary constructs containing the ubiquitin promoter driving the GUS gene and the modified hypocotyl transformation protocol coupled with hygromycin selection resulted in transformation rates of 0.4 %, 0.8% and 3.0% for B. napus RI25, B. napus BLN1239 and B. juncea 815-1-6-2 respectively. These levels were sufficient to enable experiments using the D12-desaturase cosuppression constructs to be initiated with the expectation of obtaining several independent transgenics in each species.
Selfed seed was harvested from 43 B. napus (31 of BLN1239 and 13 of RI25) and 7 B. juncea hygromycin resistant T0 plants carrying the D12-desaturase cosuppression constructs. Bulk samples of 20 seeds per plant were analysed for fatty acid composition by GC separation of fatty acid methyl esters obtained from acid-methylated oil samples (Christie, 1973). Three B. napus T0 plants (2 of BLN1239 and 1 of RI25) had oleic acid levels above 80% (control = 60-65%), while 3 lines of B. juncea had oleic acid levels above 62% (control = 40-45%).
Single seed analysis of fatty acid composition was subsequently carried out on 10 individual whole T1 seeds from each of the high-oleic T0 plants. The highest oleic acid found in single T1 seeds was 89% in B. napus and 73% in B. juncea (Figure 2). The levels of polyunsaturated fatty acids (linoleic and linolenic acid) were reduced markedly in these seeds, down to 4% in B. napus (cf. 26% in controls) and 16% in B. juncea (cf. 44% in controls). In both species the high oleic lines contained levels of saturated fatty acids (palmitic and stearic) similar to those of control lines.
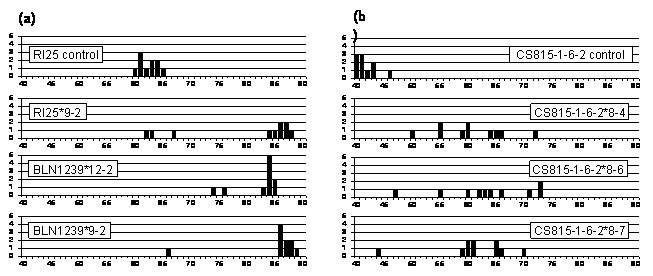 |
Figure 2: Distribution of oleic acid percentage in single seeds from control and T0 plants of (a) B. napus and (b) B. juncea
DISCUSSION
In B. napus, 43 primary (T0) transgenic plants carrying the D12-desaturase cosuppression construct have so far been analysed. Of these, 3 lines have very high levels of oleic acid, up to 89% compared to the normal level of 63%. These levels are similar to those previously reported to occur in B. napus by suppression of D12-desaturase gene activity (Kinney, 1994)
High-oleic derivatives of both B. napus lines (RI25 and BLN1239) were obtained. Southern analysis indicated that RI25*9-2 carries two copies of the transgene and BLN1239*12-2 carries six copies. The variation in oleic acid content observed between individual T1 seeds presumably reflects segregation for the transgene, and suggests that the highest oleic acid phenotype observed can be fixed in homozygous segregants isolated from the progeny of selected T1 seeds.
In B. juncea, only 7 transformants of CS815-1-6-2 have so far reached maturity. Of these, 3 lines had elevated oleic acid content, up to 73% compared to the normal level of 42%. This is the first time that these levels have been reported in B. juncea and enable this species to be immediately converted to a source of oil that meets current canola oil quality specifications.
The introduction of cosuppression constructs appears to have silenced the activity of the endogenous D12-desaturase activity in both species. In B. napus the silencing appears to be nearly complete as there are very low levels (4%) of polyunsaturated fatty acids (PUFA). PUFA levels of less than 5% were previously observed in high-oleic transgenic lines of rapeseed and soybean (Kinney, 1997). The silencing of endogenous D12-desaturase activity appears less pronounced in B. juncea, as PUFA levels are at 16% of total fatty acids. It is possible that B. juncea lines with higher levels of oleic acid will be obtained from amongst the large number of transgenic plants still to be analysed.
ACKNOWLEDGMENTS
We are very grateful for assistance from Lorraine Tonnet in the preparation and GC analysis of all oil samples.
REFERENCES
Auld, D.L, Heikkinen M.K, Erickson, D.A, Sernyk, JL and Romero J.E, 1992 Rapeseed mutants with reduced levels of polyunsaturated fatty acids and increased levels of oleic acid. Crop Science 32: 657-662
Bade, J.B. and Damm, B. (1995). Agrobacterium mediated transformation of rapeseed (Brassica napus). Pp30-38 in “Gene transfer to plants”, I. Potrykus and G. Spangenberg (eds.), Springer Lab Manual, Springer Verlag, Berlin, Heidelberg, 1995, ISBN 3-540 58406-4,
Christie, W.E. (1973) pp 88-89 in "Lipid Analysis", Pergamon, Oxford
Kinney, A.J. (1994). Genetic modification of the storage lipids of plants. Current Opinion in Biotechnology 5: 144-151
Kinney, A.J. (1997) Development of genetically engineered oilseeds: from molecular biology to agronomics. Pp 298-301 in: Physiology, Biochemistry and Molecular Biology of Plant Lipids. J.P. Williams, M.U. Khan and N. Wan Lem (eds), Kluwer Academic Press, Dordrecht, The Netherlands, 1997, ISBN 0-7923-4379-4
Singh, S.P, van der Heide, T, McKinney, S, and Green, A. (1995). Nucleotide sequence of a cDNA from Brassica juncea encoding a microsomal omega-6 desaturase. Plant Physiology 109: 1498
Wang, M.B, Upadhyaya, N.M, Brettell. R.I.S, and Waterhouse, P.M (1997). Intron-mediated improvement of a selectable marker gene for plant transformation using Agrobacterium tumifaciens. Journal of Genetics and Breeding 51: 325-334